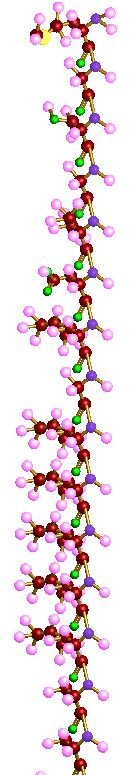
Design
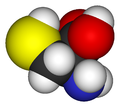
In order to model a polypeptide or protein, amino acids need to be built from
molecules containing an amine group (NH2), a carboxylic group (COOH) and a side chain
(R) as
shown below:
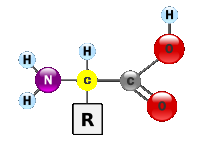
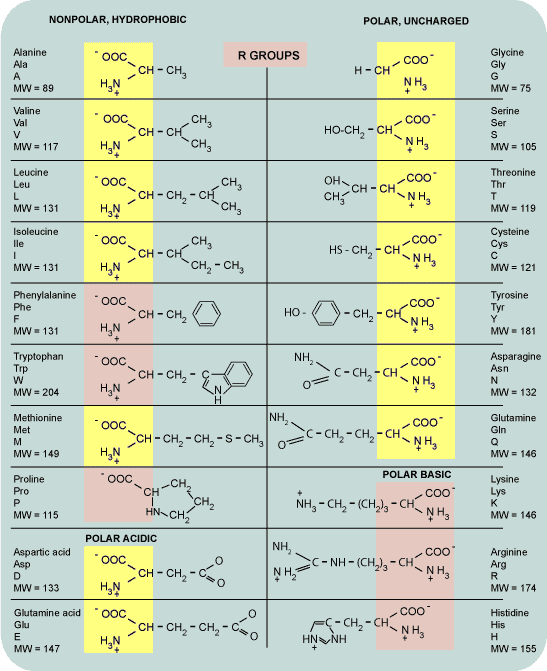
This process was accomplished by creating three main data types: Element, Bond, and Amino Acid. The elements and respective bonds were instantiated and recursively drawn starting at the central carbon atom (colored in yellow), transversing through the amine (Nitrogen and two hydrogens) group and carboxylic (Carbon and oxygens) group and then moving to the side chain (R) until the central atom was reached again. The angles between the bonds and elements were carefully calculated in order to display a 3D view of the amino acid. Once the recursive algorithm was finalized, models of the amino acids were designed and implemented. A total of 14 amino acid models were implemented and these are shown below colored in yellow:
Once the amino acid models shown above were created, the next step was to create a polypeptide, which is a group of polymers formed from the linking of amino acids. This step requiered another data type: Amino Acid data type. Links at the amine and carboxylic ends of the amino acids were broken in order for the amino acids to join together as shown below:
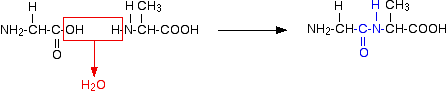
The OH and H elements were removed from the amino acids and a new bond was created in order to link both amino acids. This process continued for the entire amino acid sequence. A short sequence can be:
mfsfvdlrll lllaatallt hgqeegqveg qdedippitc vqnglryhdr dvwkpepcri
This sequence is taken from the National Center for Biotechnology Information bank, which describes the first 60 amino acids that apha-1 type I collagen (COL1A1) polypeptide contains.
Once the entire polypeptide is built, it can then be rendered as a linear group of "electron shells" as shown below for Cysteine amino acid or shown in the "Screenshot" section for a full polypeptide or protein.
In order to model a protein, multiple polypeptides fold, twist and come together by means of hydrogen bonding. As they come together, they entangle and form the final structure known as a protein. This was implemented by joining polypeptide data types in order to form a protein. Screenshots of a protein is shown in the "Screenshot" section.
An overview of the entire process is shown below:
Different materials were applied to each of the elements and lighting and animation was provided to model the sequence of steps required to form a protein.
References
1. National Center for Biotechnology Information (NCBI): http://www.ncbi.nlm.nih.gov/
2. GLUI user interface Library: http://glui.sourceforge.net/#download
3. Wikipedia Encyclopedia for general information about the amino acids. For example, Cysteine: http://en.wikipedia.org/wiki/Cysteine
4. The structure of proteins: http://www.chemguide.co.uk/organicprops/aminoacids/proteinstruct.html