Project Description
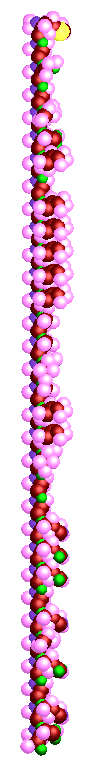
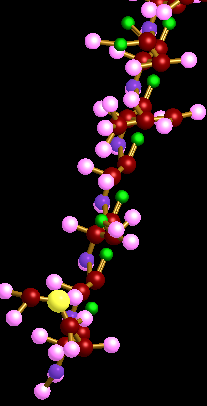
3D Protein simulation is a simulating tool designed to model amino acids, polypeptides and a protein. The process starts at the element level structure by bonding elements to form amino acids, joining amino acids to form polypeptides, and then combining polypeptides to form a protein.In order to simulate this process, the simulation is divided into three main sections to allow the user to render fourteen out of twenty amino acids, render up to ten polypeptides, and render a single protein. The polypeptides and protein rendering depends on the user's input file (.p or .pp), which contains a list of amino acids that form that particular polypeptide or protein.
A graphical user interface was created to ease the process of selecting the various options which include:
1. Selecting different rendering modes for polypeptides and proteins . These include rendering the structure as a linear electron group of shells or as a planar framework structure.
2. Animating a 3D structure by rotating, moving, or zooming in or out
3. Managing the lighting sources to either change the light or move the light around the rendered structure
4. A legend that shows the elements coloring code as well as information of the chosen amino acid
5. Selecting different amino acids to render (14 out of 20) in order to study their 3D structure
6. Allow the flexibility to control the length of the polypeptide chain or protein
All these capabilities combined allows the user to interact, learn, and study these structures in much greater detail.